what will happen to the unattached trna once it has delivered its amino acid
How does the cell catechumen DNA into working proteins? The process of translation can be seen as the decoding of instructions for making proteins, involving mRNA in transcription as well equally tRNA.
The genes in Deoxyribonucleic acid encode protein molecules, which are the "workhorses" of the jail cell, carrying out all the functions necessary for life. For example, enzymes, including those that metabolize nutrients and synthesize new cellular constituents, as well as DNA polymerases and other enzymes that brand copies of DNA during prison cell partitioning, are all proteins.
In the simplest sense, expressing a gene ways manufacturing its respective protein, and this multilayered process has two major steps. In the offset stride, the data in DNA is transferred to a messenger RNA (mRNA) molecule past manner of a process chosen transcription. During transcription, the DNA of a gene serves as a template for complementary base of operations-pairing, and an enzyme chosen RNA polymerase II catalyzes the formation of a pre-mRNA molecule, which is and so candy to course mature mRNA (Figure 1). The resulting mRNA is a unmarried-stranded copy of the gene, which next must exist translated into a poly peptide molecule.
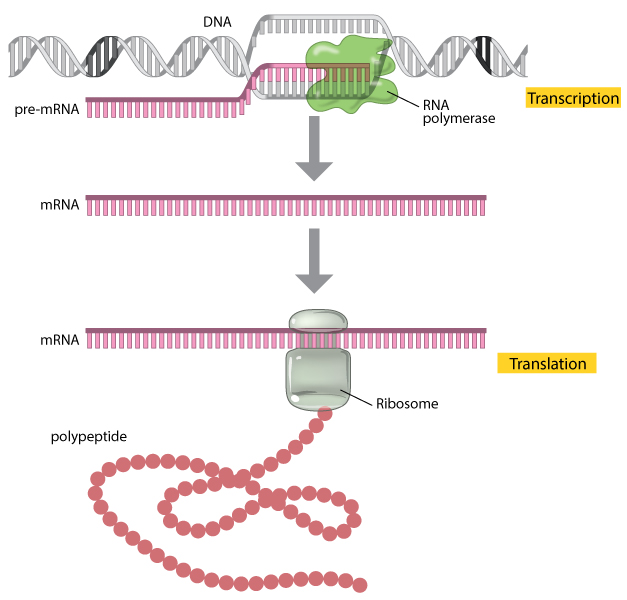
Figure 1: A factor is expressed through the processes of transcription and translation.
During transcription, the enzyme RNA polymerase (dark-green) uses DNA as a template to produce a pre-mRNA transcript (pink). The pre-mRNA is processed to course a mature mRNA molecule that can exist translated to build the poly peptide molecule (polypeptide) encoded past the original gene.
During translation, which is the 2nd major step in gene expression, the mRNA is "read" according to the genetic lawmaking, which relates the DNA sequence to the amino acid sequence in proteins (Figure 2). Each group of three bases in mRNA constitutes a codon, and each codon specifies a particular amino acrid (hence, it is a triplet code). The mRNA sequence is thus used every bit a template to gather—in order—the chain of amino acids that grade a poly peptide.
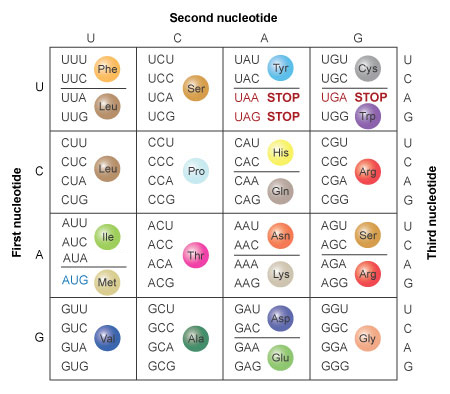
Figure two: The amino acids specified past each mRNA codon. Multiple codons can lawmaking for the same amino acid.
The codons are written 5' to 3', as they announced in the mRNA. AUG is an initiation codon; UAA, UAG, and UGA are termination (terminate) codons.
But where does translation have place within a prison cell? What individual substeps are a part of this process? And does translation differ between prokaryotes and eukaryotes? The answers to questions such equally these reveal a dandy deal well-nigh the essential similarities betwixt all species.
Where Translation Occurs
Within all cells, the translation machinery resides within a specialized organelle called the ribosome. In eukaryotes, mature mRNA molecules must leave the nucleus and travel to the cytoplasm, where the ribosomes are located. On the other hand, in prokaryotic organisms, ribosomes can attach to mRNA while it is still existence transcribed. In this situation, translation begins at the five' end of the mRNA while the three' finish is still attached to Deoxyribonucleic acid.
In all types of cells, the ribosome is composed of two subunits: the large (50S) subunit and the modest (30S) subunit (S, for svedberg unit, is a measure of sedimentation velocity and, therefore, mass). Each subunit exists separately in the cytoplasm, but the two join together on the mRNA molecule. The ribosomal subunits contain proteins and specialized RNA molecules—specifically, ribosomal RNA (rRNA) and transfer RNA (tRNA). The tRNA molecules are adaptor molecules—they take one end that can read the triplet code in the mRNA through complementary base of operations-pairing, and another finish that attaches to a specific amino acrid (Chapeville et al., 1962; Grunberger et al., 1969). The thought that tRNA was an adaptor molecule was kickoff proposed by Francis Crick, co-discoverer of DNA structure, who did much of the central work in deciphering the genetic lawmaking (Crick, 1958).
Within the ribosome, the mRNA and aminoacyl-tRNA complexes are held together closely, which facilitates base-pairing. The rRNA catalyzes the attachment of each new amino acid to the growing chain.
The Offset of mRNA Is Not Translated
Interestingly, non all regions of an mRNA molecule represent to particular amino acids. In particular, there is an surface area near the 5' stop of the molecule that is known as the untranslated region (UTR) or leader sequence. This portion of mRNA is located between the starting time nucleotide that is transcribed and the commencement codon (AUG) of the coding region, and information technology does not affect the sequence of amino acids in a protein (Figure 3).
Then, what is the purpose of the UTR? It turns out that the leader sequence is important considering it contains a ribosome-binding site. In leaner, this site is known as the Shine-Dalgarno box (AGGAGG), later on scientists John Smoothen and Lynn Dalgarno, who first characterized it. A similar site in vertebrates was characterized by Marilyn Kozak and is thus known every bit the Kozak box. In bacterial mRNA, the 5' UTR is normally brusk; in human mRNA, the median length of the 5' UTR is about 170 nucleotides. If the leader is long, it may incorporate regulatory sequences, including binding sites for proteins, that tin can affect the stability of the mRNA or the efficiency of its translation.
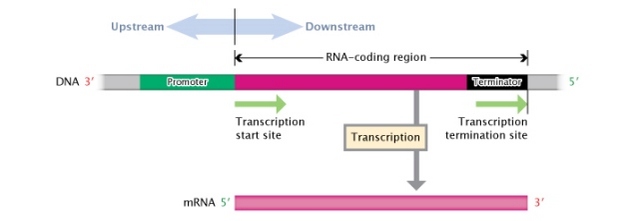
Effigy three: A DNA transcription unit.
A Dna transcription unit is composed, from its 3' to v' end, of an RNA-coding region (pinkish rectangle) flanked by a promoter region (green rectangle) and a terminator region (blackness rectangle). Regions to the left, or moving towards the three' end, of the transcription start site are considered \"upstream;\" regions to the right, or moving towards the 5' cease, of the transcription start site are considered \"downstream.\"
© 2014 Nature Educational activity Adapted from Pierce, Benjamin. Genetics: A Conceptual Approach, 2d ed. All rights reserved. ![]()
Translation Begins Subsequently the Assembly of a Complex Structure
The translation of mRNA begins with the germination of a complex on the mRNA (Figure iv). First, three initiation cistron proteins (known as IF1, IF2, and IF3) bind to the small-scale subunit of the ribosome. This preinitiation complex and a methionine-conveying tRNA then bind to the mRNA, almost the AUG beginning codon, forming the initiation complex.
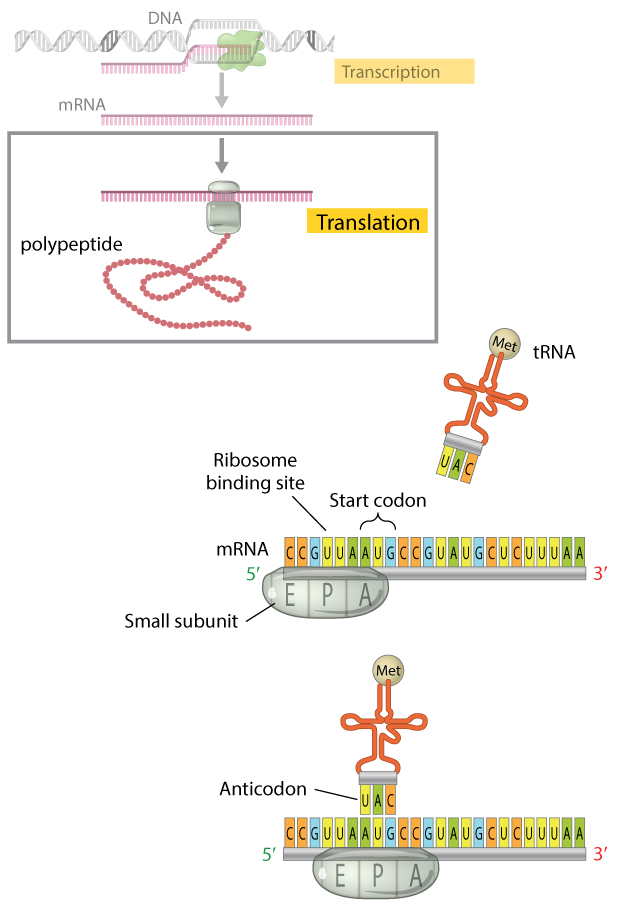
Figure four: The translation initiation complex.
When translation begins, the minor subunit of the ribosome and an initiator tRNA molecule gather on the mRNA transcript. The small subunit of the ribosome has three binding sites: an amino acid site (A), a polypeptide site (P), and an exit site (E). The initiator tRNA molecule conveying the amino acid methionine binds to the AUG start codon of the mRNA transcript at the ribosome'due south P site where it will go the offset amino acid incorporated into the growing polypeptide chain. Here, the initiator tRNA molecule is shown binding subsequently the small ribosomal subunit has assembled on the mRNA; the order in which this occurs is unique to prokaryotic cells. In eukaryotes, the gratuitous initiator tRNA first binds the pocket-size ribosomal subunit to form a complex. The complex then binds the mRNA transcript, so that the tRNA and the small ribosomal subunit bind the mRNA simultaneously.
Although methionine (Met) is the first amino acrid incorporated into any new protein, information technology is not e'er the starting time amino acid in mature proteins—in many proteins, methionine is removed later on translation. In fact, if a large number of proteins are sequenced and compared with their known cistron sequences, methionine (or formylmethionine) occurs at the N-terminus of all of them. However, non all amino acids are equally likely to occur 2nd in the chain, and the 2nd amino acid influences whether the initial methionine is enzymatically removed. For case, many proteins brainstorm with methionine followed by alanine. In both prokaryotes and eukaryotes, these proteins have the methionine removed, so that alanine becomes the N-terminal amino acid (Table i). However, if the second amino acid is lysine, which is besides frequently the instance, methionine is not removed (at least in the sample proteins that have been studied thus far). These proteins therefore begin with methionine followed past lysine (Flinta et al., 1986).
Table 1 shows the N-terminal sequences of proteins in prokaryotes and eukaryotes, based on a sample of 170 prokaryotic and 120 eukaryotic proteins (Flinta et al., 1986). In the table, M represents methionine, A represents alanine, G represents lysine, Southward represents serine, and T represents threonine.
Tabular array 1: N-Terminal Sequences of Proteins
| North-Terminal Sequence | Percent of Prokaryotic Proteins with This Sequence | Percent of Eukaryotic Proteins with This Sequence |
| MA* | 28.24% | 19.17% |
| MK** | 10.59% | 2.50% |
| MS* | 9.41% | xi.67% |
| MT* | 7.65% | 6.67% |
* Methionine was removed in all of these proteins
** Methionine was not removed from whatever of these proteins
Once the initiation complex is formed on the mRNA, the large ribosomal subunit binds to this complex, which causes the release of IFs (initiation factors). The large subunit of the ribosome has 3 sites at which tRNA molecules can demark. The A (amino acrid) site is the location at which the aminoacyl-tRNA anticodon base pairs upwards with the mRNA codon, ensuring that correct amino acid is added to the growing polypeptide chain. The P (polypeptide) site is the location at which the amino acid is transferred from its tRNA to the growing polypeptide chain. Finally, the E (leave) site is the location at which the "empty" tRNA sits before being released dorsum into the cytoplasm to demark another amino acrid and repeat the procedure. The initiator methionine tRNA is the merely aminoacyl-tRNA that can bind in the P site of the ribosome, and the A site is aligned with the second mRNA codon. The ribosome is thus ready to bind the second aminoacyl-tRNA at the A site, which will be joined to the initiator methionine past the first peptide bond (Figure five).

Figure 5: The large ribosomal subunit binds to the small ribosomal subunit to complete the initiation circuitous.
The initiator tRNA molecule, carrying the methionine amino acid that volition serve as the commencement amino acrid of the polypeptide chain, is spring to the P site on the ribosome. The A site is aligned with the side by side codon, which volition be bound past the anticodon of the next incoming tRNA.
The Elongation Phase
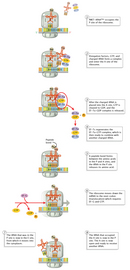
The side by side phase in translation is known as the elongation stage (Effigy 6). First, the ribosome moves forth the mRNA in the v'-to-3'direction, which requires the elongation factor G, in a process called translocation. The tRNA that corresponds to the 2d codon tin then demark to the A site, a footstep that requires elongation factors (in East. coli, these are called EF-Tu and EF-Ts), as well as guanosine triphosphate (GTP) as an energy source for the procedure. Upon binding of the tRNA-amino acid complex in the A site, GTP is broken to class guanosine diphosphate (Gdp), then released along with EF-Tu to be recycled by EF-Ts for the next round.
Side by side, peptide bonds between the now-next get-go and second amino acids are formed through a peptidyl transferase action. For many years, information technology was thought that an enzyme catalyzed this footstep, only recent evidence indicates that the transferase action is a catalytic function of rRNA (Pierce, 2000). Afterward the peptide bail is formed, the ribosome shifts, or translocates, again, thus causing the tRNA to occupy the E site. The tRNA is then released to the cytoplasm to pick up some other amino acid. In addition, the A site is now empty and set up to receive the tRNA for the adjacent codon.
This process is repeated until all the codons in the mRNA have been read by tRNA molecules, and the amino acids attached to the tRNAs accept been linked together in the growing polypeptide chain in the appropriate club. At this point, translation must be terminated, and the nascent protein must be released from the mRNA and ribosome.
Termination of Translation
There are 3 termination codons that are employed at the end of a poly peptide-coding sequence in mRNA: UAA, UAG, and UGA. No tRNAs recognize these codons. Thus, in the identify of these tRNAs, one of several proteins, called release factors, binds and facilitates release of the mRNA from the ribosome and subsequent dissociation of the ribosome.
Comparing Eukaryotic and Prokaryotic Translation
The translation process is very similar in prokaryotes and eukaryotes. Although different elongation, initiation, and termination factors are used, the genetic code is mostly identical. Equally previously noted, in bacteria, transcription and translation take place simultaneously, and mRNAs are relatively short-lived. In eukaryotes, however, mRNAs accept highly variable half-lives, are subject to modifications, and must get out the nucleus to exist translated; these multiple steps offer boosted opportunities to regulate levels of protein production, and thereby fine-tune factor expression.
References and Recommended Reading
Chapeville, F., et al. On the role of soluble ribonucleic acrid in coding for amino acids. Proceedings of the National Academy of Sciences 48, 1086–1092 (1962)
Crick, F. On poly peptide synthesis. Symposia of the Lodge for Experimental Biology 12, 138–163 (1958)
Flinta, C., et al. Sequence determinants of N-last protein processing. European Journal of Biochemistry 154, 193–196 (1986)
Grunberger, D., et al. Codon recognition by enzymatically mischarged valine transfer ribonucleic acid. Scientific discipline 166, 1635–1637 (1969) doi:10.1126/science.166.3913.1635
Kozak, M. Point mutations close to the AUG initiator codon affect the efficiency of translation of rat preproinsulin in vivo. Nature 308, 241–246 (1984) doi:ten.1038308241a0 (link to article)
---. Point mutations ascertain a sequence flanking the AUG initiator codon that modulates translation past eukaryotic ribosomes. Cell 44, 283–292 (1986)
---. An analysis of v'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Research 15, 8125–8148 (1987)
Pierce, B. A. Genetics: A conceptual approach (New York, Freeman, 2000)
Polish, J., & Dalgarno, L. Determinant of cistron specificity in bacterial ribosomes. Nature 254, 34–38 (1975) doi:10.1038/254034a0 (link to article)
Source: https://www.nature.com/scitable/topicpage/translation-dna-to-mrna-to-protein-393/
0 Response to "what will happen to the unattached trna once it has delivered its amino acid"
Post a Comment